Grow in the Dark: Systems Biology of Hormone-Regulated Etiolated Seedling Growth
We study cross-regulation of gene expression by hormone signaling pathways during seedling growth.
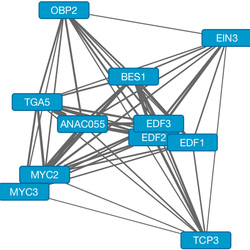
The phenotype of an organism results from a vast number of information inputs that change over time. Inputs come from both internal and external factors, such as weather conditions, pathogen attack and the activity of hormone signaling pathways. Dynamic regulation of transcription is a key point at which inputs are integrated and processed to determine phenotypic outputs. This occurs through changes in the activity and binding of TFs that may regulate gene expression both positively and negatively. An individual TF binds to the promoters of hundreds or thousands of genes, acting as an information input to the expression of those genes. Tens of TFs may be bound simultaneously to the promoter of any given gene. The final transcriptional output of each gene is therefore determined by a complex interaction between many different TFs. Fluctuations in expression of that gene reflect changes in the population of bound TFs. The major hormone signaling pathways interact in complex ways, both synergistic and antagonistic. Each pathway regulates dynamically a suite of TFs that affect expression of downstream genes. These pathways influence growth and development of plants throughout their lifecycle. The result is a highly complex network of interactions that change over time. We study the interactions between hormone signaling pathways using genome-scale techniques (chromatin immunoprecipitation sequencing - ChIP-seq) to measure binding of TFs, differential gene expression (RNA-seq) and associated protein activity (proteomics, phosphoproteomics). Our targets are the major plant hormone signaling pathways, including jasmonic acid, ethylene and salicylic acid. Data are integrated using bioinformatic and systems biology techniques with the ultimate goal of generating models describing seedling growth. The is a collaboration with Joe Ecker at the Salk Institute. |
Ready, Set, Grow: Transcriptional Networks Regulating Seed Germination
Germination is a critical step in plant growth. No other developmental program will proceed if it is not completed successfully. Nutrient resources and responses to the environment vary dramatically between seeds of different plant species. However, the core theme of germination is the occurrence of a dynamic developmental process following the correct environmental stimuli. This involves extensive transcriptional reprogramming. We are taking a systems biology approach to investigate changes in transcriptional networks during seed germination, applying high-throughput sequencing technologies. Most recently we have established single-cell and tissue-specific sequencing methods to enable us to understand how gene regulatory networks specify the functions of different cell-types.
A systems-level understanding of the transcriptional networks that regulate germination will greatly enhance our understanding of plant growth and development. We relate TFs to specific transcriptional programs and to observable phenotypes using dynamic modeling approaches. Moreover, we relate interspecific differences in germination to variation in transcriptional regulation to better understand the biology of different seed types. |
Plants on Film: High-Throughput Plant Phenomics
Plants are surprisingly dynamic. Across a day their leaves move up-and-down and side-to-side. Their stems can circle around very deliberately, looking for something to grab hold of. These movements occur slowly compared to the movements of animals, so need to be looked at in a different way. Our lab uses time-lapse imaging to analyze the movements of hundreds plants at once, measuring features down to sub-millimetre scale. We have a particular interest in the development of new image analysis algorithms that improve the speed, convenience and accuracy of analyzing very large collections of images. This work is collaborative with Eddie Custovic, Jim Whelan and Dennis Deng at La Trobe.
|
The ARC Research Hub for Medicinal Agriculture
As members of the ARC MedAg Hub we work in the lab and the field on optimizing performance of medicinal plants. These projects combine both fundamental and applied biology, particularly the application of genomics to understand biochemical pathway regulation and genetic diversity in natural populations.
|
La Trobe Genomics Platform
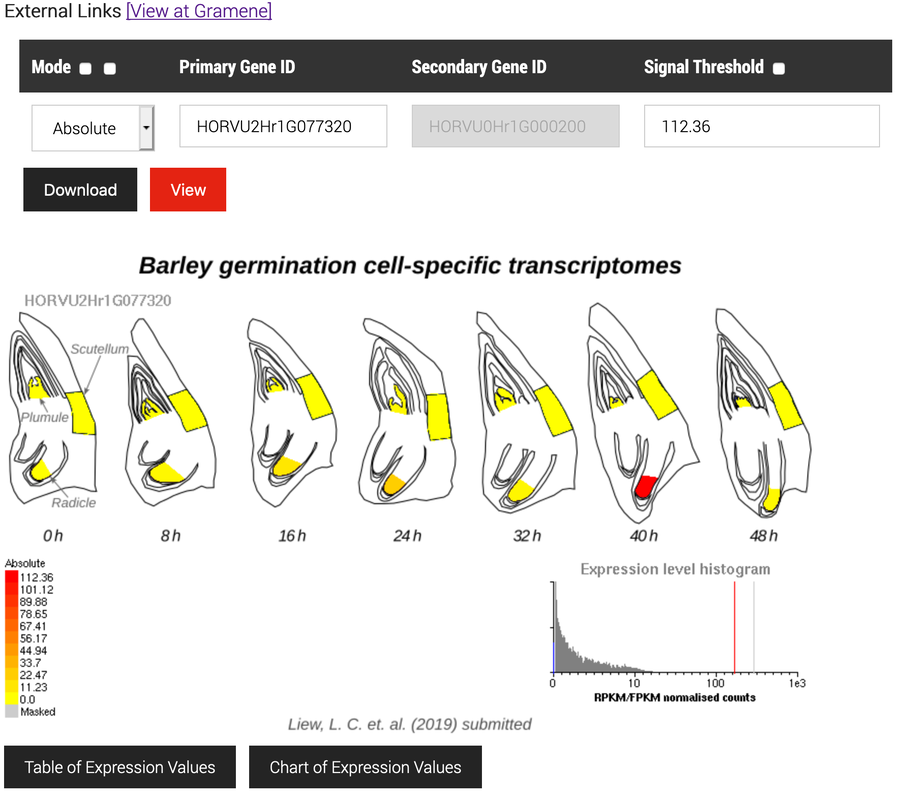
The researchers of the La Trobe Genomics Platform are embedded within our lab. The dedicated laboratory and bioinformatic staff provide an end-to-end service for both internal and external researchers. Areas of specialty include transcriptomics, single-cell analyses (using the 10x Chromium Platform), meta-analyses and machine learning. Our clients span plant, animal and medical research. We also specialize in customized public display of data, for example https://expression.latrobe.edu.au/efp/agriseqdb/barley/liew2019.
|